ACDC Challenge
Deep Learning
In this project, my teamate and I completed an automated pipeline to tackle the ACDC using Deep
Learning. Including, data prepossessing,
segmentation, features extraction, and classification. (Pytorch)
Learn more on ACDC
Segmentation
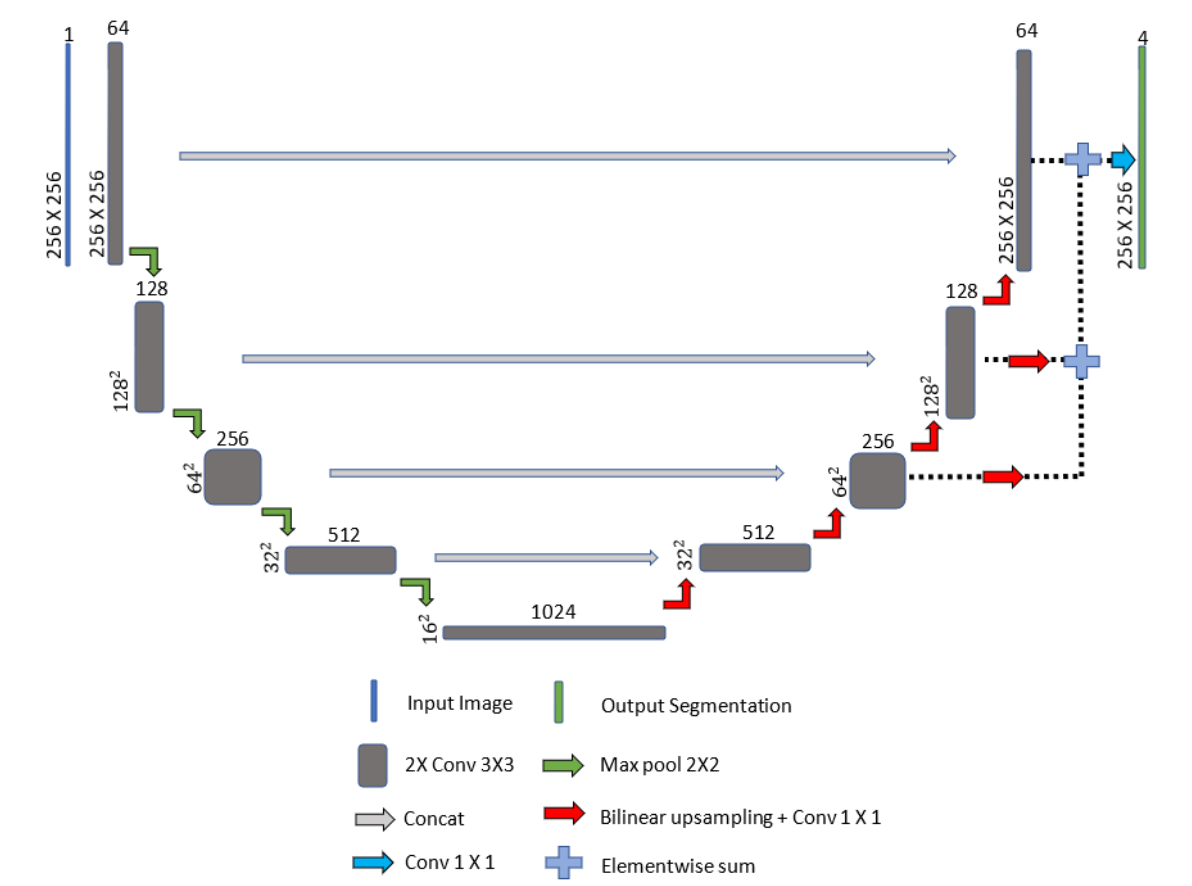
We implemented the traditional Unet first[1], and several modifications were made.
Bilinear upsampling followed by a one by one convolution layer was used instead of
transposed convolution to avoid checkerboard-like
pattern of varying magnitudes when processing 2D images.
To speed up the convergence, improve the discriminative ability of our
network, and alleviate class imbalance. We used Deep Supervision (element-wise sum and upsampling)
to combine segmentation maps at different resolutions to a matching resolution.
Classification
Both dynamic and instant features were extracted. The features were selected to quantify the traditional clinical assessment procedures by describing static and dynamic properties. These features were used to train an ensemble of MLP and a random forest for pathology classification. The softmax outputs of all MLPs were averaged and are combined sequentially with the random forest output to obtain the final ensemble prediction.
Results
We achieve dice scores of 0.945(LV), 0.885(RV), 0.90(LVM) on the ACDC test set, which earned us first place in the segmentation competetion of the project. Our Result also ranked TOP 10 on the leader-board.
[1] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Con- volutional networks for biomedical image segmenta- tion,” in International Conference on Medical image computing and computer-assisted intervention, pp. 234– 241, Springer, 2015.